Spatial Information
Bioelectric Morphogenesis Projects in the Levin Lab
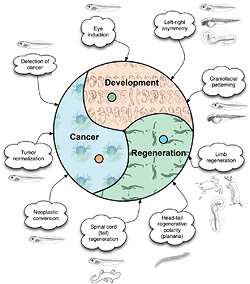
- Cancer as a disease of geometry
- Detection of cancer via bioelectric signatures
- Neoplastic conversion without genetic damage
- Tumor normalization by enlarging computational boundary of cells
- Development
- Crarniofacial patterning
- Morphogenesis of new synthetic organisms
- Left-right asymmetry scaling from cytoskeleton to whole body axis
- Regeneration
- Limb regeneration in amphibians and mammals
- Head-tail regenerative polarity (planaria)
- Planarian shape control and species' attractors in morphospace
- Spinal cord (tail) regeneration
Summary
The capacity to generate a complex organism from the single cell of a fertilized egg is one of the most amazing qualities of multicellular creatures. The processes involved in laying out a basic body plan and defining the structures that will ultimately be formed depend upon a constant flow of information between cells and tissues. The Levin laboratory studies the molecular mechanisms that cells use to communicate with one another in the 4-dimensional dynamical system known as the developing embryo. We also study the flow of information necessary for an injured system to recognize what structures must be rebuilt, and the algorithms that coordinate individual cell activity towards specific patterning outcomes during remodeling and cancer suppression. Through experimental approaches and mathematical modeling, we examine the processes governing large-scale pattern formation and biological information storage during animal embryogenesis and regeneration. Our investigations are directed toward understanding the mechanisms of signaling between cells and tissues that allows a biological system to reliably generate, maintain, and repair a complex morphology as a kind of navigation of morphospace. We study these processes in the context of embryonic development, regeneration and cancer, with a particular emphasis on the biophysics of cell behavior. In complement to other groups focusing on gene expression networks and biochemical signaling factors, we are pursuing, at a molecular level, the roles of endogenous voltages, and bioelectric gradients as epigenetic carriers of morphological information and as a kind of "cognitive glue" that binds individual cells toward common purpose in their activity. Using gain- and loss-of-function techniques to specifically modulate cells' ion flow we have the ability to regulate large-scale morphogenetic events relevant to limb formation, eye induction, craniofacial and neural patterning, limb regeneration, head remodeling, etc. While our focus is on the fundamental mechanisms of pattern regulation, this information will also result in important clinical advances through harnessing bioelectrical controls of cell behavior for regenerative medicine. Our goal is to understand how to use low-complexity triggers (morphogenetic subroutine calls) to kickstart, not micromanage, a complex repair cascade. An example is the 24-hour bioreactor treatment of an adult frog leg which causes a year and a half of autonomous regeneration.
Bioelectrical controls of vertebrate appendage regeneration

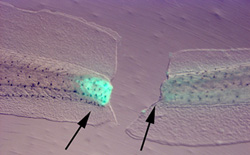

Regeneration is a fascinating example of pattern regulation, and has important biomedical implications. A regenerating system must not only recognize damage, but also pursue a goal-directed process of restoring the missing structures (and crucially, know to stop when this process is complete, thus avoiding cancerous overgrowth). Interestingly, systems with high regenerative ability have low susceptibility to neoplasm, contrary to the simple view in which cellular plasticity and propensity for proliferation should go together in cancer and regeneration. Instead, data suggest that the morphogenetic controls imposed during regeneration can prevent cells from ignoring the patterning cues of the host (as occurs in many cancers). What is the mechanistic nature of these controls? Our lab studies the role of voltage gradients, and how these biophysical controls couple to genetic and epigenetic pathways in the induction of regeneration and the imposition of correct morphology on the restored tissue. We mainly use two model systems to understand these processes: Xenopus laevis tadpoles and planarian flatworms. An important part of our mission is the development of molecular reagents, protocols, databases, transgenic animals, and conceptual (modeling) tools to facilitate others' study of bioelectrical signals in many aspects of morphogenesis in diverse model systems.
While vertebrate regeneration is considered to be limited, the Xenopus tadpole is able to regenerate its tail - a complex appendage containing spinal neurons, muscle, skin, and vasculature. We identified three electrogenic proteins whose activity is required for the production of a depolarization zone that underlies regeneration in the blastema and demonstrated that a proton flux from the wound epithelium is necessary and sufficient to drive the downstream events of regeneration, including cell proliferation, innervation, and expression of regeneration-specific markers. We are currently working on inducing regeneration of limbs, eyes, tails, and craniofacial structures in normally non-regenerating species by providing the appropriate bioelectric signals to the cells at the wound site. While ion flows control cell-level behaviors such as migration, differentiation, and proliferation, bioelectric signals also function as master regulators of large-scale shape in many contexts: a simple signal can induce complex, highly orchestrated, self-limiting downstream morphogenetic cascades. For example, an unmodulated flux of protons can cause the formation of a complete tail of the right rise and tissue composition. Inducing the host to form structures it already knows how to make is a very desirable property for regenerative medicine approaches since it avoids the inevitable complexity explosion of having to micromanage (directly bioengineer) the creation of complex organs and appendages. We are also pursuing novel long-range sources of patterning information for regeneration, such as the body-wide bioelectric cues that regulate formation of the nascent brain and suppression of tumorigenesis.
Bioelectrical, non-local controls of regenerative polarity in planaria
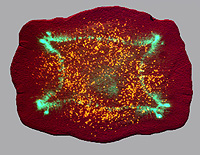
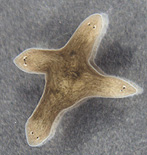
Planarian flatworms have an impressive capacity for regeneration. They are able to regenerate large parts of the body, and are continuously maintained by a well-characterized resident population of adult stem cells. Upon cutting, these organisms are able to regenerate the head and tail at their appropriate locations. What mechanisms determine the polarity and allow tissue re-patterning to take place? Consider: after bisection, cells on one side of the cut (in the head fragment's posterior end) will form a tail, while cells which were their immediate neighbors before the cut will make a head (the tail fragment's anterior half). Our data suggest that the mechanism by which blastema cells poll the rest of the host (to determine where the wound is located and what other tissues already exist in the fragment and thus don't need to be recreated) is mediated by physiological signals passing through nerves and long-range gap junctional paths. We have identified endogenous ion fluxes and voltage gradients maintained by specific ion pumps which are crucial for the determination of anterior-posterior polarity during regeneration; manipulating these signals allows us to specify tissue identity and thus control the anatomical structure of regenerating worms. Through studying the roles of electrical polarity (maintained by ion channel and gap junction systems) in planarian regeneration we are gaining insight into the control of regeneration and morphogenesis by endogenous ion fluxes and into the mechanisms by which stem cell differentiation is integrated into functional organ/tissue systems within the organism. Most importantly, we've recently shown that a rapid, transient alteration of the physiological signals underlying morphostasis and regeneration is maintained in perpetuity! That is, worms forming 2 heads (one at each end) because of a 2-day disruption of gap junctional signals will continue to form 2 heads through subsequent months of amputation or fission in normal conditions. These data illustrate how information embedded in physiological networks can be solidified into permanent alteration of the large-scale structure bodyplan. More broadly, this work identifies a molecular glimpse of how the "target morphology" of an animal (the form towards which regeneration regulates) can be permanently reset, and reveals that a drastic change in body structure and behavior can be maintained across a complex metazoan organism's normal mode of reproduction without any change in DNA sequence.
Cancer as a problem of morphogenetic disorganization
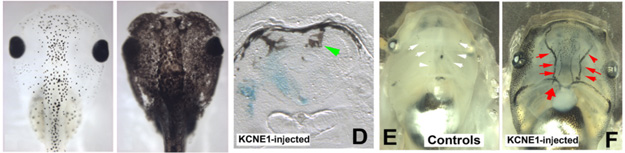
One view of cancer (distinct from the current paradigm of intrinsically "cancerous" cells resulting from specific DNA modifications) is as a problem of organization within a "society of cells". Cancer is, in some sense, a disease of geometry - a failure of cells to attend to the signals that normally organize their behavior towards the patterning needs of the host. This view is supported by classical and recent data showing that aggressive cancer cells can be normalized by regenerative and embryonic environments - context is crucial, and environments in which tight morphogenetic cues are being imposed have the ability to reverse or reboot cancer phenotypes. The converse is also true: we have found a bioelectrical property that imposes a neoplastic-like phenotype upon pigment cells in vivo. This is not surprising, given that a significant component of morphogenetic cues are ionic in nature. Remarkably however, this effect is non-local in nature - it is the transmembrane potential of other, quite distant cells that determines the metastasis-like effect. We are pursuing several lines of inquiry including 1) developing methods for tracking bioelectrical signatures of pre-tumor cells as a non-invasive diagnostic modality, 2) understanding how voltage properties of distant cells can be a cancer-triggering event in the body, and 3) learning to control transmembrane potential of key cell types to normalize existing tumors.
Left-Right Asymmetry
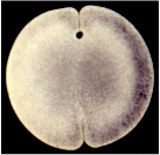
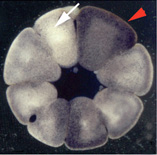
The vertebrate body plan is basically bilaterally-symmetrical;
however, consistent and well-conserved asymmetries of the brain and
visceral organs are superimposed upon the fundamental structure.
Asymmetries in the left-right axis present a number of deep puzzles
which link evolutionary biology, clinical medicine, biochemistry,
embryology, cognitive science, and perhaps even quantum parity
violations. Strikingly, it is now known that even single mammalian
cells in culture maintain a consistent left-right axis. We are
working to understand the mechanisms by which the embryo aligns the
left-right axis with respect to the other two axes, and imposes this
spatial information on macroscopic cell fields prior to the
morphogenesis of the asymmetric organs. In
contrast
to popular models of asymmetry initiation by extracellular fluid
flow during gastrulation, our lab studies much earlier,
intracellular events that break symmetry and establish consistent asymmetry as a form of
planar cell polarity, using physiological mechanisms to amplify
cytoskeletal chirality
across cell fields.
Gap Junctions in Pattern Formation
While asymmetrically expressed genes have been identified in several vertebrate systems, many critical questions remain about how cellular polarity is synchronized and amplified across embryonic fields to allow cells to ascertain their position with respect to the midline. We identified a dependence of asymmetric gene expression on early communication between left and right sides in the chick and frog. For example, expression of left-sided markers depends on events occurring on the right side, during very early stages, suggesting that the two sides need to coordinate their decision with respect to the L-R identity of each. One mechanism for communicating between cells and tissues involves gap junctions: multimers of connexin proteins form channels between cells and pass small molecules, subject to complex regulation by various signals. Using misexpression of Connexin proteins and their mutants to disrupt and induce long-range gap junctional paths in early chick and frog embryos, we showed that gap junctions are crucially involved in L-R patterning in early embryos of Xenopus and chick. The data suggest the presence of a unidirectional circumferential flow of small molecules through gap junctions across the whole embryonic field during blastula/early gastrula stages. Our research focuses on understanding the mechanisms upstream and downstream of specific gap junction communications (GJC) in embryos, as they relate to pattern formation and growth control, and on identifying the small molecule morphogens that traverse junctions. More broadly, we focus on gap junctions as a bioelectric patterning element that sets up domains of isopotential cell fields during morphogenesis. More recently, we have identified roles for the electrical synapses known as gap junctions in mediating bioelectric regulation of tumorigenesis, brain patterning, and neural pathfinding.
Bioelectric Aspects of Very Early Left-Right Patterning
L-R asymmetry can only be derived from gap-junctional movement of determinants that is directionally biased. In frog and chick embryos, we have identified a set of four ion transporters which reliably establish a voltage gradient along the midline. This gradient is required for normal asymmetry, and our current quantitative models suggest that it is sufficient to redistribute small molecule morphogens from left to right through the gap junctional paths. These transporters establish a battery across the midline, and in Xenopus, this occurs by the second cell cleavage. Molecular localization of ion channel/pump proteins, and direct detection of asymmetric ion flows (H+ and K+ ions) reveal that these embryos know their left from their right within about 2 hours of fertilization. What establishes the right-sided ion flow? Our latest work has focused on the cytoskeleton, and we showed that the early protein localization machinery is consistently right-biased, allowing intracellular kinesin and dynein motors to deliver maternal ion transporter cargo to the right ventral blastomere (thus establishing the battery and electromotive force for the trans-junctional morphogen(s)). We are currently characterizing the intracellular microtubule organizing center whose chirality is likely to be the ultimate origin of asymmetry, as its orientation with the other two axes is established during fertilization. Remarkably, the role of cytoskeletal proteins in directing asymmetry is conserved even across independent origins of multicellularity, as we were able to show that the same tubulin mutations randomize asymmetry in frog, human cells, and C. elegans as do in plants.We have also pursued roles for planar cell polarity in spreading LR information across a blastoderm.
The Role of Serotonin in Embryogenesis
The importance of serotonin in neuronal function is well established. Interestingly, it also has roles in early embryogenesis, long before nerve systems appear. This is probably indicative of evolutionarily early systems of cell signaling which became co-opted by neurons when they arose. Taking advantage of the well-characterized pharmacology and genetics of many steps in the serotonin signaling pathway, we are studying how serotonin signaling is used in information exchange between cells in processes such as L-R patterning and control of timing and cell movement during gastrulation. We have shown that serotonin is utilized by both chick and frog embryos, at very early stages, as a small molecule signal which is transported in a left-right gradient and regulates the development of laterality. Indeed, we now know that the early frog embryo is literally an electrophoresis chamber, which uses voltage potentials to generate consistently biased left-right gradients in serotonin in an epigenetic process not dependent on zygotic gene expression. We have modeled this process quantitatively, and characterized novel intracellular serotonin-binding proteins which directly activate asymmetric gene expression after their rightward movement, linking an early biophysical process to transcriptional regulation via chromatin modification pathways. Serotonin is also a key mediator for bioelectric control of neuronal outgrowth from transplants
Mathematical Modeling and Physiomics
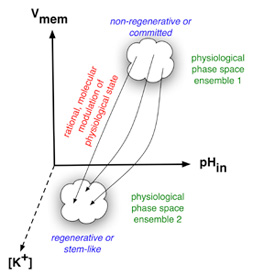
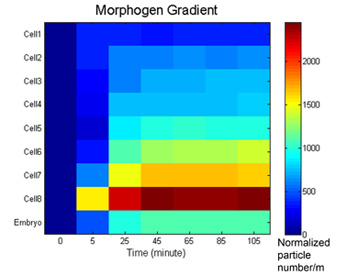
Molecular biology and genomics are revealing a constantly expanding amount of information about genes, their products, and the way they interact. It is notoriously difficult to control or make predictions about systems involving mutual interactions of even a few components because of feedback loops and the basic results of dynamical systems theory. Indeed, looking at a high-resolution mechanistic pathway, such as painstakingly elucidated in many recent studies, is insufficient for knowing what biological pattern this transcriptional network results in. It is essential to develop constructive, synthetic models of morphogenesis which integrate 3-dimensional shape from the function of molecular components and pathways. To fully understand the implications of information coming from genome projects and biochemical analyses of gene activities for morphogenesis, a synthesis is needed. We are attempting to use the mathematical and computer modeling tools of chaos, information, and complexity theories to understand large-scale patterning and control properties of bioelectrical mechanisms and small molecule transport among cell groups. Our main efforts along these lines are directed towards (1) development of a formalization for morphogenetic processes (bioinformatics of shape, beyond gene/protein sequence, and automated model discovery), (2) testing the hypothesis that cell behavior can be understood as the segregation and movement of cell states through a multi-dimensional state space with axes defined by bioelectrical parameters such as membrane voltage, K+ content, pH, nuclear membrane potential, etc., and (3) developing quantitative models integrating physiology and genetics of ion transporter function during early left-right asymmetry.
Computational Approaches to Pattern Formation:
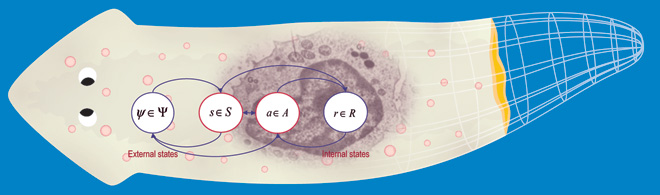

Bioinformatics has revolutionized molecular biology, but the field still largely lacks necessary computational tools to crack the problem of pattern regulation and control. With each new functional or high-resolution genomic dataset, it becomes ever more difficult to come up with constructive models that explain how complex pattern arises, and what steps could be taken to induce the kinds of patterning changes required for regenerative medicine and the repair of birth defects. Thus, we are working towards a set of software tools that utilize the principles of machine learning and artificial intelligence to establish a bioinformatics of shape. Our goal is to understand the fundamental rules that allow complex patterns to be formed and to repair themselves after damage, and to discover models of these processes that are not merely lists of necessary gene products but algorithms that clearly reveal the regulation of shape, size, topology, and anatomical arrangement. To this end, we have produced novel formalisms for describing functional experiments and resulting anatomical outcomes, and a genetic algorithm-based platform for discovery of mechanistic models that explain complex patterning results in the published literature. This resulted in the first quantitative model of planarian regeneration explaining numerous experimental datasets, and the first regenerative model discovered by a non-human intelligence (a machine learning platform). Importantly, our goal is not only to understand specifics of real model organisms (e.g., planarian regeneration) but to uncover broad general principles useful for synthetic morphology applications - an Artificial Life perspective. Another aspect of this work is the study of computation in biological tissues (which process information in order to regulate and remodel their shape). Thus for example, we are not simply trying to make computational models to explain planarian regeneration, but rather, we see the planarian itself as a model of the kind of computation we want to understand. Additional projects include computational models of emergent signaling dynamics that explain stochastic behavior and pattern dysregulation in cancer, and the use of computational models to understand how bioelectric circuits and transcriptional machinery interact to make decisions about growth and form.


