Educational Materials
Joel Grodstein: Computational Bioelectricity (EE 123)
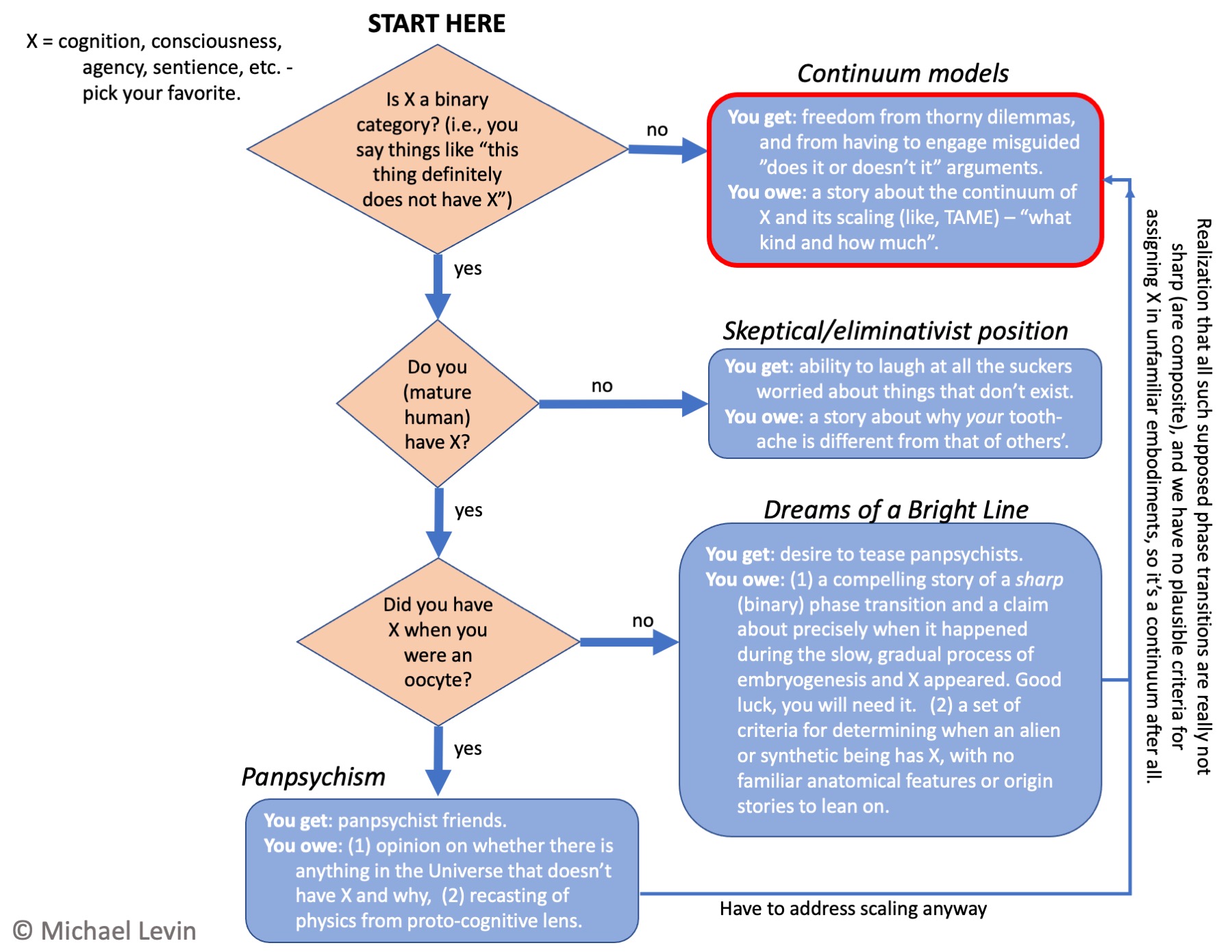
Decision chart for views on cognition
Biological software and hardware
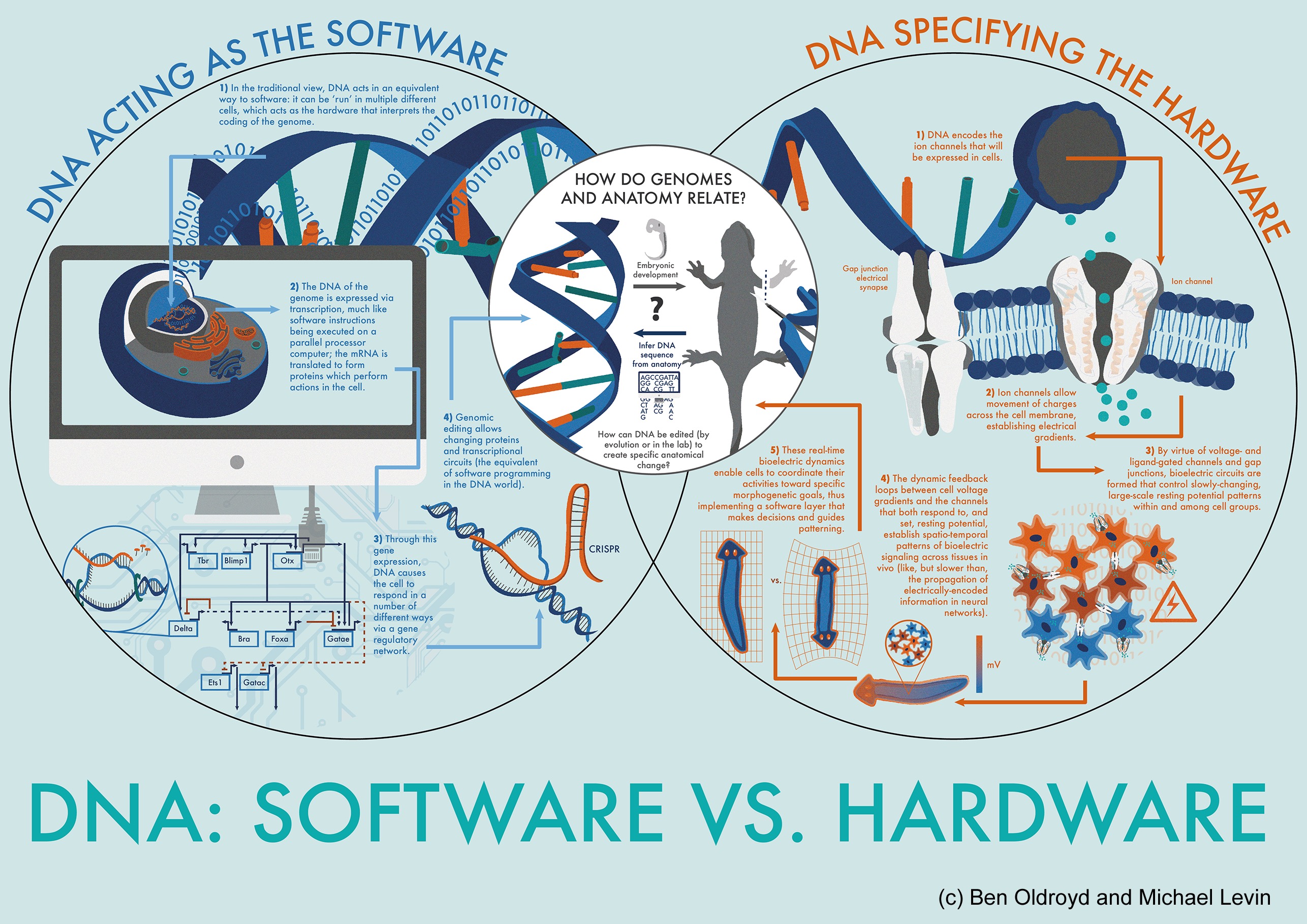
Hardware-software
Multi-scale pattern control (during regulative development, metamorphosis, cancer suppression, and regenerative repair) requires living systems to predictably reach invariant goal states from a diverse range of starting conditions and perturbing influences. This is implemented by a set of computational processes in which cells and tissues need to make decisions about current anatomical state and stop remodeling when the correct target morphology has been reached. Computational systems can be described via two components: the software (a set of dynamic processes including communication among subunits, information processing, and memory) and the hardware (the physical components whose dynamical interactions implement the software). A traditional view sees the cell as the hardware that interprets DNA sequences as the software. A different view is that the genome encodes the hardware, providing each cell with versatile computational elements such as ion channels. The software in this case is the electrophysiological dynamical system formed by coupled cells. Such bioelectrical networks can form among all cell types, although they are best known in the brain. Much as the ion channel and synaptic hardware allow the brain to implement numerous kinds of computational behaviors (e.g., goal-seeking and memory circuits), the same hardware in other cell types is a much more ancient system which tissues use to execute the computations needed for anatomical control (pattern memory and global integration of cells toward the correct complex outcome).
Multi-modal anatomical control
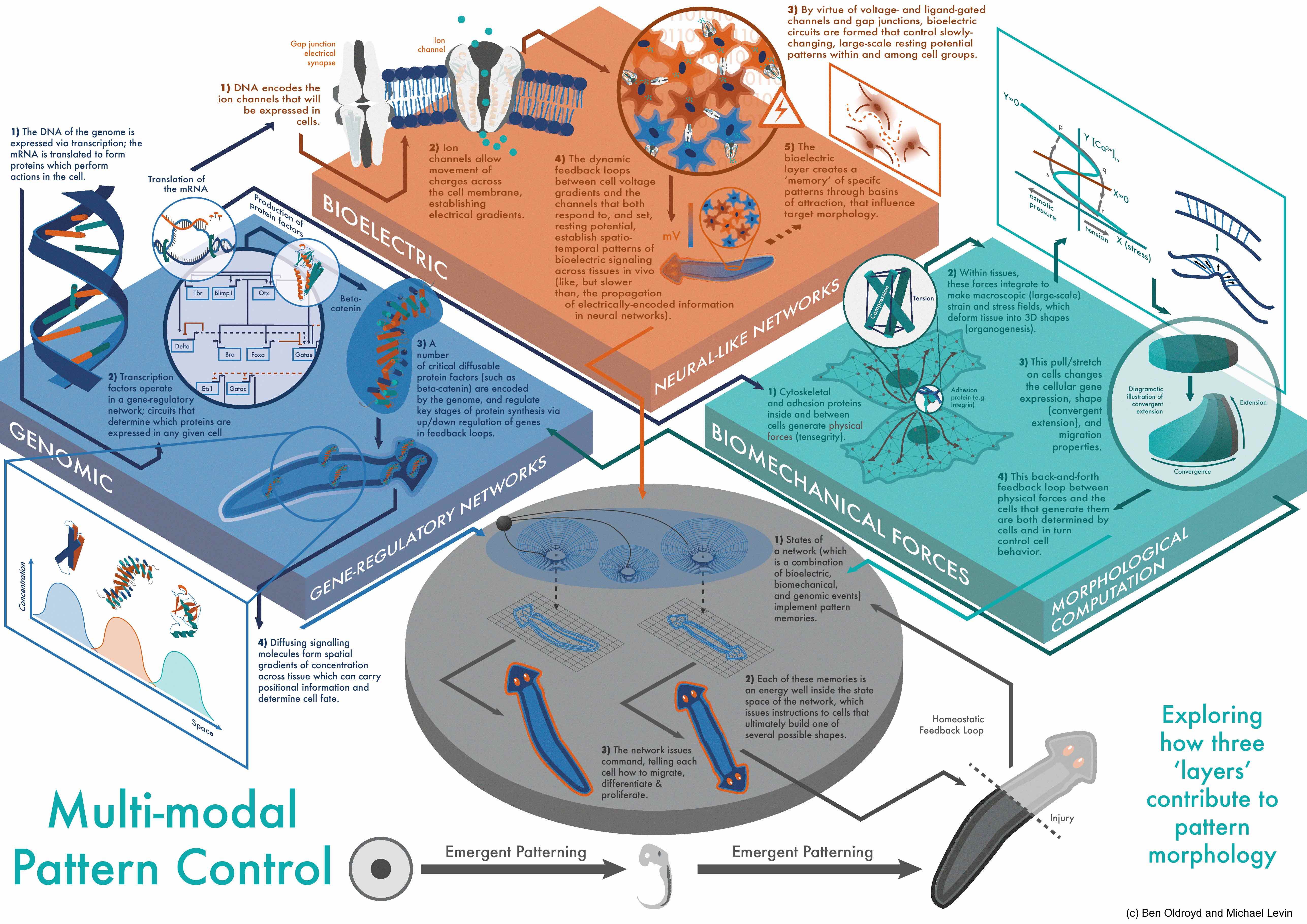
3-layers
Dynamic control of complex anatomical pattern is a multi-scale process that occurs during embryogenesis, regeneration, metamorphosis, remodeling, and cancer suppression. Individual cells and cell networks must coordinate their activity toward specific tissue- and organ-level morphological outcomes. These processes take place through three integrated layers: 1) gene-regulatory networks comprised of transcriptional circuits and diffusible biochemical signals, 2) physical forces – biomechanical processes by which tensions and stresses implement morphological computation, and 3) bioelectric networks – neural-like dynamics in gap junction-coupled tissues expressing ion channels and pumps. Each of these layers processes information and implements emergent complexity at cell, tissue, organ, and whole body axis levels. They are coupled, with bi-directional functional links between each, and unique roles in decision-making and implementation of pattern control. In this way, physics, chemistry, and computer science interplay to enable biological systems to regulate their own structure and function with a high degree of adaptive robustness.
These illustrations were made by Ben Oldroyd, via support of the Templeton World Charity Foundation.